Brief description of the project
Project title: IRN AP14871383 «Development of KASP markers to facilitate construction of competitive bread wheat, durum wheat, and barley cultivars in Kazakhstan».
Relevance. Kazakhstan is an important exporter of cereals in the world market. Bread wheat, durum wheat and barley, occupying 12.2, 0.5, and 2.0 million hectares of sown area, respectively, are the main cereal crops in the country. Cereal grains are an important commodity on the international market, and the country is in the top ten exporters. However, the average yield rarely exceeds 1 t/ha for wheat and 1.5 t/ha for barley. Due to the widespread use of genomic technologies, the use of SNP (single nucleotide polymorphism) markers and SNP-associated types of DNA markers can significantly help to the development of new highly productive competitive cultivars. One of the most common type of markers is KASP (Kompetitive-allele-specific PCR), which provides fast, reliable, cheap and automated screening of genotypes. The development of KASP markers is based on SNPs previously identified in genome-wide association studies for these crops. The use of KASP sets will increase accuracy and reduce the time required for new cultivars breeding.
The goal of the project: to develop sets of effective KASP markers for bread wheat, durum wheat and barley for the breeding of new competitive cultivars with high yield and grain quality in the main grain-growing regions of the country, as well as to introduce a marker-assisted selection method into the breeding process.
Expected results:
- SNP markers associated with adaptation traits, yield components, disease resistance and grain quality in three crops will be transformed into KASP markers.
- Commercial cultivars and promising lines of bread wheat, durum wheat and barley will be genotyped using KASP markers.
- Validation of the effectiveness of KASP markers will be carried out using available field data for three crops from the northern, central and southeastern regions of the country.
- KASP sets for three crops will be evaluated; practical recommendations will be prepared to improve breeding processes using marker-assisted breeding methods.
- Based on the results of the project, an information database will be developed for commercial cultivars and promising lines of bread wheat, durum wheat and barley.
Scientific Supervisor of the project:
Turuspekov Yerlan, Head of the Laboratory of Molecular Genetics, Candidate of Biological Sciences, Professor. Co-author of over 100 publications, incl. 50 in peer-reviewed international publications indexed in Web of Science databases (H-index 13, Researcher ID: C-3458-2011) and/or with a CiteScore percentile in the Scopus database (H-index 16, Scopus Author ID: 57197860996), including 29 articles for the period 2016-2021. ORCID https://orcid.org/0000-0001-8590-1745.
Research group:
- Sereda, Candidate of Agricultural Sciences, Head of the Department of breeding and primary seed production at the A. Khristenko Karaganda breeding station, breeding experience >35 years. Co-author of 10 cultivars of grain crops. H-index=4 (Scopus Author ID: 57210671318).
- Chudinov, Head of the Barley Breeding Department at the Karabalyk breeding station. H-index 6 (Scopus Author ID: 55600618100), is the main co-author of 10 cultivars of spring barley, 1 cultivar of oats and 4 cultivars of spring bread wheat, co-author of 62 scientific articles, holder of 10 patents for breeding achievements.
- Sh. Anuarbek, Researcher, PhD, has more than 20 scientific papers, incl. 6 articles (5 in peer-reviewed journals, WoS and Scopus (H-index 3, Scopus Author ID: 57192177334); 3 articles in COXON journals), 2 patents (utility model), 1 scientific and methodological recommendations, etc. ORCID https:/ /orcid.org/0000-0002-8673-9820
- Amalova, Researcher, PhD-student, co-author of 5 articles (2 WoS, H-index 3, ResearcherID: AAO-1156-2020), Scopus Author ID: 57213623570; 3 CCSES). ORCID https://orcid.org/0000-0002-7903-3467
- Genievskaya, MSc, Junior researcher, PhD-student on the project topic, co-author of 11 articles (9 in peer-reviewed journals, WoS and Scopus), (H-index 5, Scopus Author ID: 57196939730), 7 of them on the project topic. Co-author of a patent (utility model). Trained at the University of Oxford, UK.
Foreign Consulting Scientist: Professor Simon Griffiths, John Innes Centre, Norwich, UK. Has >100 articles indexed in the Web of Science and Scopus databases (H-index 34 Scopus Author ID: 7102341547).
List of publications of the project’s participants (2015-2020)
- Zatybekov, A. Genievskaya Y, Rsaliyev A, Turuspekov Y, Abugalieva S. (2022). Identification of quantitative trait loci for leaf rust and stem rust seedling resistance in bread wheat using a genome-wide association study. Plants (IF=3.935, quartile-Q1; SJR=0.892, percentile-56) 11(1):74. DOI: 10.3390/plants11010074.
- Amalova, A., Abugalieva S, Babkenov A., Babkenova S., Turuspekov Y. (2021). Genome-wide association study of yield components in spring wheat collection harvested under two water regimes in Northern Kazakhstan. PeerJ (IF=3.369, quartile-Q2; SJR=0.927, percentile-83) 9:e11857. DOI:10.7717/peerj.11857.
- Yermekbayev K., Griffiths S., Chhetry M., Leverington-Waite, M., Orford, S., Amalova, A., Abugalieva S., Turuspekov Y. (2020). Construction of a Genetic Map of RILs Derived from Wheat ( aestivum L.) Varieties Pamyati Azieva × Paragon Using High-Throughput SNP Genotyping Platform KASP-Kompetitive Allele Specific PCR. Russian Journal of Genetics (IF=0.729, quartile-Q4, SJR=0.208, percentile-8) 56(9):1090-1098. DOI: 10.1134/S102279542009015X.
- Anuarbek S, Abugalieva S, Pecchioni N, Laidò G, Maccaferri M, Tuberosa R, Turuspekov Y. (2020) Quantitative trait loci for agronomic traits in tetraploid wheat for enhancing grain yield in Kazakhstan environments. PLoS ONE (IF=3.788, quartile-Q2; SJR=0.990, percentile-92). 15(6): e0234863. DOI: 10.1371/journal.pone.0234863.
- Genievskaya Y, Turuspekov Y, Rsaliyev A, Abugalieva S. (2020). Genome-wide association mapping for resistance to leaf, stem, and yellow rusts of common wheat under field conditions of South Kazakhstan. PeerJ (IF=3.369, quartile-Q2; SJR=0.927, percentile-83) 8:e9820. DOI: 10.7717/peerj.9820.
- Almerekova S., Sariev B., Abugalieva A., Chudinov V., Sereda G., Tokhetova L., Ortaev A., Tsygankov V., Blake T., Chao S., Genievskaya Y., Abugalieva S., Turuspekov Y. (2019). Association mapping for agronomic traits in six-rowed spring barley from the USA harvested in Kazakhstan, PLoS ONE (IF=3.788, quartile-Q2; SJR=0.990, percentile-92). 14(8):e0221064. DOI: 10.1371/journal.pone.0221064.
- Genievskaya Y., Almerekova S., Sariev B., Chudinov V., Tokhetova L., Sereda G., Ortaev A., Tsygankov V., Blake T., Chao S., Sato K., Abugalieva S., Turuspekov Y.(2018). Marker-trait associations in two-rowed spring barley accessions from Kazakhstan and the USA. PLoS ONE (IF=3.788, quartile-Q2; SJR=0.990, percentile-92) 3(10): e0205421. DOI: 10.1371/journal.pone.0205421.
- Turuspekov Y., Plieske J., Ganal M., Akhunov E., Abugalieva S. (2017) Phylogenetic analysis of wheat cultivars in Kazakhstan based on the wheat 90K single nucleotide polymorphism array. Plant Genetic Resources: Characterisation and Utilisation (IF=1.243, quartile-Q4, SJR=0.343, percentile-62). 15(1):29-35.DOI: 10.1017/S1479262115000325.
- Turuspekov Y., Baibulatova A., Yermekbayev K., Tokhetova L., Chudinov V., Sereda G., Ganal M.W, Griffiths S., Abugalieva S. (2017). GWAS for plant growth stages and yield components in spring wheat (Triticum aestivum) harvested in three regions of Kazakhstan. BMC Plant Biology (IF=4.96, quartile-Q1; SJR=1.378, percentile-87). 17(S1):51-61. DOI: 10.1186/s12870-017-1131-2
- Tajibayev D., Yusov V.S., Chudinov V.A., Mal’chikov P.N., Rozova M.A., Shamanin P., Shepelev S., Sharmag R., Tsygankov V.I., Morgounov A. I. (2021). Genotype by environment interactions for spring durum wheat in Kazakhstan and Russia. Ecological Genetics and Genomics (SJR=0.990, percentile-74), 21, 100099. DOI: 10.1016/j.egg.2021.100099
Results for 2022:
- The DNA of samples of the collection of cultivars and promising lines were isolated and purified. The DNA-bank of the collection of bread wheat (30), durum wheat (10), barley (20) has been replenished.
- As a result of the identification of quantitative trait loci (QTL) using genome-wide association study and QTL analysis in mapping populations, SNP markers were selected for the transformation into KASP markers.
- Using the nucleotide sequences of SNP markers associated with plant adaptation traits, yield components, disease resistance and grain quality, primers were designed for bread wheat (34 KASP markers), durum wheat (15 KASP markers) and barley (12 KASP markers).
Results for 2023:
- DNA of a collection of cultivars and promising lines of kernels crops was isolated and purified. The DNA bank of a collection of bread wheat ( 100 ), durum wheat ( 41 ), barley ( 100 ) was replenished.
- As a result of identification of quantitative trait loci ( Quantitative Trait Loci – QTL, QCL) based on genome-wide association analysis study (GWAS) and QTL analysis of mapping populations, SNP markers were selected for transformation into KASP type markers. Based on the nucleotide sequences of the isolated SNP markers associated with plant adaptation traits, yield components, disease resistance and grain quality, primers were designed for bread wheat (88 KASP), durum wheat (15 KASP) and barley ( 46 KASP).
- Genotyping of commercial varieties and promising lines of grain crops was carried out using KASP markers: soft wheat – 60 markers, hard wheat – 25 markers, barley – 50 markers.
Publications 2023
List of publications in peer-reviewed scientific journals indexed in the Web of Science and (or) Scopus databases
- Genievskaya Y., Almerekova S., Abugalieva S., Abugalieva A., Sato K., Turuspekov Y. Identification of SNPs associated with grain quality traits in spring barley collection grown in southeastern Kazakhstan // Agronomy( IF= 3.7, Q1 Agronomy; SJR= 0.668, percentile: 84 — Agricultural and Biological Sciences, Agricultural and Biological Sciences ). – 2023. – Vol. 13(8). – P. 1560 https://doi.org/10.3390/agronomy13061560 [in English].
Abstracts of the international conference (indicating the form of the report ):
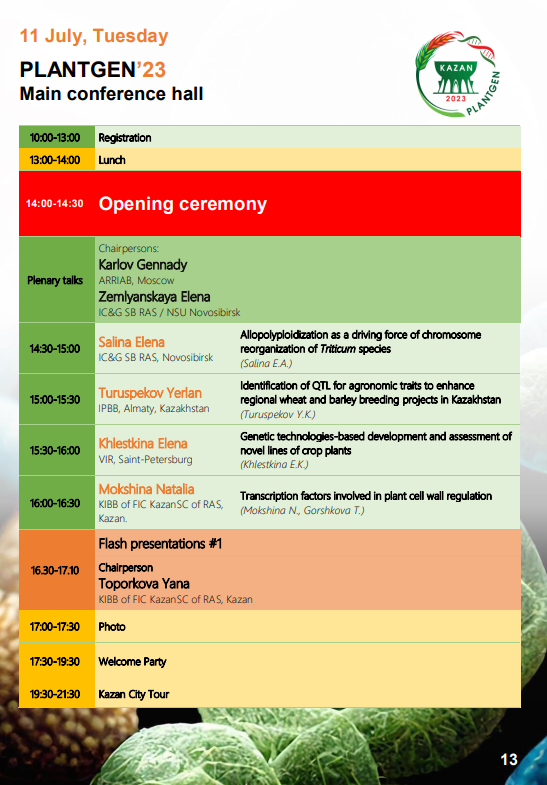
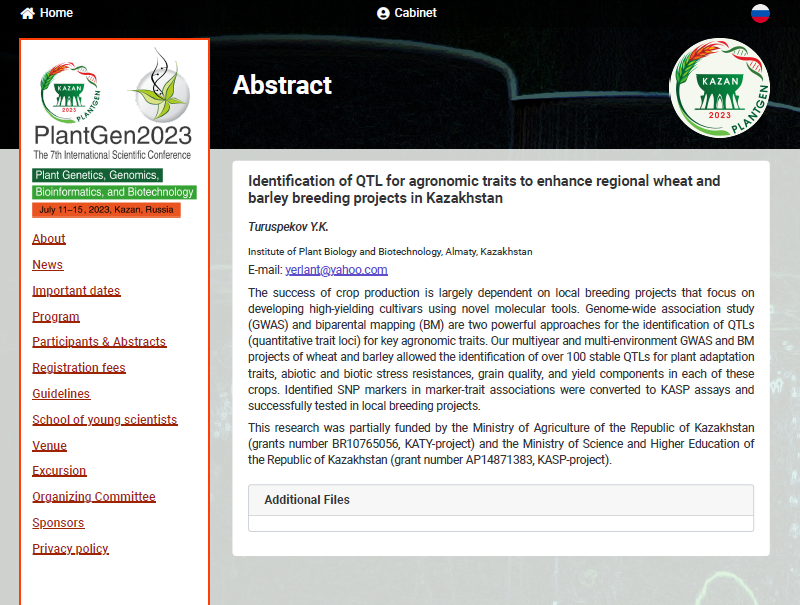
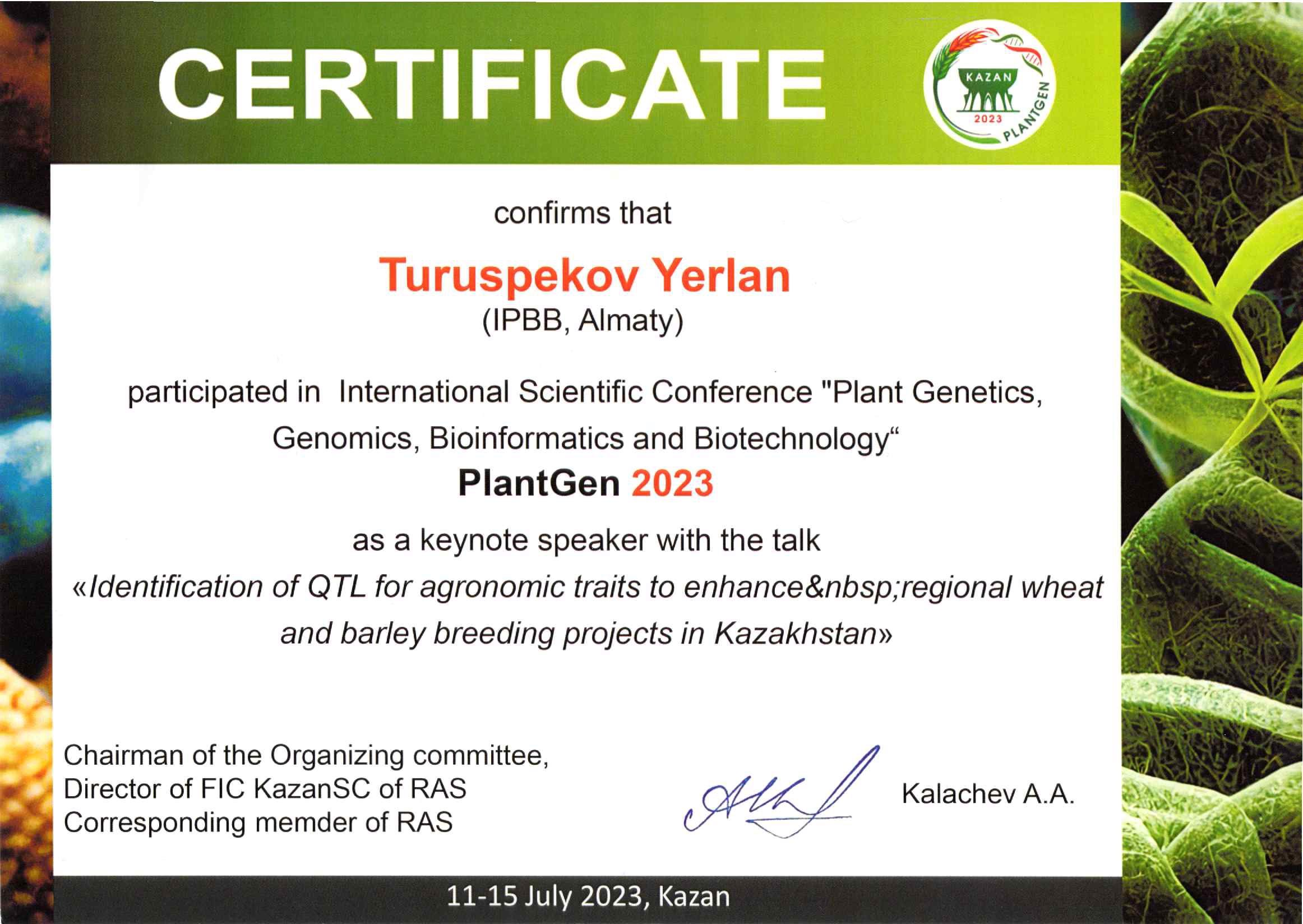
- Turuspekov Y. Identification of QTL for agronomic traits to enhance regional wheat and barley breeding projects in Kazakhstan // The 7th international conference PlantGen20231, Kazan, June 10-15, 2023. (plenary report). [in English]
Results for 2024:
- DNA of a collection of cultivars and promising lines of kernels crops was isolated and purified. The DNA bank of a collection of bread wheat (30), durum wheat (21), barley (40) was replenished.
- As a result of identification of quantitative trait loci (Quantitative Trait Loci – QTL, QCL) based on genome-wide association analysis study (GWAS) and QTL analysis of mapping populations, SNP markers were selected for transformation into KASP type markers. Based on the nucleotide sequences of the isolated SNP markers associated with plant adaptation traits, yield components, disease resistance and grain quality, primers were designed for bread wheat (78 KASP), durum wheat (20 KASP) and barley ( 42 KASP).
- Phenotyping of commercial varieties and promising lines of cereal crops was carried out in three regions of the country. KASP markers were assessed and polymorphic markers were identified for soft wheat (161), durum wheat (37) and barley (76). Sets of KASP markers for key agronomic traits were created for three regions.
- An information database for cereal crops was developed based on the results of phenotyping and genotyping.
Publications 2024
List of publications in peer-reviewed scientific journals indexed in the Web of Science and (or) Scopus databases
- Amalova A., Babkenov A., Philp C., Griffiths S., Abugalieva S., & Turuspekov, Y . Identification of quantitative trait loci associated with plant adaptation traits using nested association mapping population // Plants ( IF= 3.7, Q1 Agronomy; SJR= 0.795, percentile: 73 -Plant Science). – 2024. – Vol.13(18). – P. 2623. https://doi.org/10.3390/plants13182623 [in English]
Articles in a peer-reviewed domestic publication with a non-zero impact factor (recommended by the SC CQASHE RK):
- Anuarbek S., Chudinov V., Sereda G., Babkenov A., Savin T., Fedorenko E., Tsygankov V., Tsygankov A., Amalova A., Turuspekov Y. Yield stability analysis of bread wheat genotypes in Kazakhstan // Bulletin of KazNU . Biological Series. — 2024. — Vol 3 (100). — P 56-68. https://doi.org/10.26577/bb.2024.v100.i3.05 [in English]
Abstracts of the international conference (indicating the form of the report ):
- Anuarbek S., Pecchioni N., Laidò G., Maccaferri M., Tuberosa R., Abugalieva S., Turuspekov Y. Quantitative trait places for agronomic traits in tetraploid wheat field-tested in Kazakhstan environments // « International Conference on Plant Biology and Biotechnology (ICPBB 2024) »: Proceedings of the international conference / edited by E.K. Turuspekov , S.I. Abugalieva . – Almaty: IBBR, 2024 – 66 p. (oral report) ) [in English]
- Amalova A., Yessimbekova M., Ortaev A. , , Turuspekov Y. Griffiths S., Burakhoja A., Abugalieva S. Genome-wide association study of agronomic traits in winter wheat collection grown in Kazakhstan //“International Conference on Plant Biology and Biotechnology (ICPBB 2024)»: Proceedings of the international conference / edited by E.K. Turuspekov , S.I. Abugalieva . — Almaty: IBBR, 2024 — 73 p. (oral report) [in English]
- Amalova A., Babkenov A., Philp C., Griffiths S., Abugalieva S., Turupekov Y. Genome-wide association study of plant adaptation traits in nested association mapping population grown in Kazakhstan / /“ International Conference on Plant Biology and Biotechnology (ICPBB 2024)»: Proceedings of the international conference / edited by E.K. Turuspekov , S.I. Abugalieva . — Almaty: IBBR, 2024 — 87 p. (poster) [in English]
- Anuarbek Sh ., Genievskaya Y., Zatybekov A., Pecchioni N., Laidò G., Rsaliyev A., Chudinov V., Turuspekov Y., Abugalieva S. Genome-wide association study of leaf rust and stem rust seedling and adult resistances in tetraploid wheat accessions harvested in Kazakhstan / /“ International Conference on Plant Biology and Biotechnology (ICPBB 2024)»: Proceedings of the international conference / edited by E.K. Turuspekov , S.I. Abugalieva . — Almaty: IBBR, 2024 — 88 p. (poster) [in English]
Patent for utility model:
- Genievskaya Yu.A., Turuspekov E.K., Abugalieva S.I. «A method for identifying highly productive lines of barley ( Hordeum vulgare ) using KASP genotyping technology» // National Institute of Intellectual Property of the Ministry of Justice of the Republic of Kazakhstan. Electronic bulletin. — 2024. Application number — 2024/0537. [in Russian]
Monograph
- Abugalieva S.I. , Amalova A.Y., Turuspekov E.K. “Identification of quantitative trait loci for bread wheat productivity based on the use of biparental mapping populations.” – Almaty: Piramida Group, 2024. – 210 p [in Russian].