Brief description of the project
(2022-2024)
Project title: IRN AP14870612 «Study of the genetic diversity in species of the genus Tulipa L. growing in Kazakhstan».
Relevance. Tulipa L. is a group of perennial geophyte monocots, one of the largest genera of the Liliaceae family. It has a great economic, floricultural, ecological importance, cultural value in many regions of the world. Most species are found in mountainous areas, 17 species grow in the steppes and adjacent deserts and semi-deserts. Most of the species are subjected to anthropogenic impact, but 18 species are included in the Red Book of Kazakhstan. The use of modern molecular genetic and genomic technologies with the involvement of bioinformatics, along with traditional botanical methods, help to refine the taxonomy and study the phylogenetic and evolutionary relationships of wild plant species.
The goal of the project: To study intra- and interspecific genetic diversity of wild-growing species of tulips in Kazakhstan using SSR markers, to determine the phylogenetic relationships of species by nucleotide sequences of DNA barcoding markers and chloroplast genome using new generation genomic technologies, as well as phytochemical traits of widespread tulip species.
Expected results:
- Populations of tulip species will be collected, species identity will be determined, herbariums will be compiled and digitized.
- Phylogenetic relationships of Tulipa species will be studied using the genetic diversity of chloroplast genome.
- Phylogenetic relationships of representatives of the genus Tulipa will be analyzed using markers of the nuclear and chloroplast genome.
- The level of intra- and interspecific genetic diversity of Tulipa species of Kazakhstan will be studied, the population structure will be determined.
- Widespread species of tulips will be studied for secondary metabolites.
- The database «Biodiversity of the flora of Kazakhstan» will be supplemented with data on botanical collections, molecular genetic data of deposited nucleotide sequences of chloroplast genomes and DNA barcoding data of species of the genus Tulipa.
Scientific Supervisor of the project: S. Abugalieva, principal researcher, Doctor of Biological Sciences, Professor. In 2017-2022 co-authored 40 scientific articles, including 29 articles in peer-reviewed foreign scientific journals indexed in Web of Science (https://www.webofscience.com/wos/author/record/2577530) and/or Scopus (H-index=8, https://www.scopus.com/authid/detail.uri?authorId=6507029042), 11 in journals recommended by the Committee for Control in the Sphere of Education and Science (CCSES) of the Ministry of Education and Science of the Republic of Kazakhstan.
Research group:
- Turuspekov, Candidate of biological sciences, Professor, Head of the Laboratory of Molecular Genetics. He has >100 scientific publications, including 40 in peer-reviewed international journals, indexed in Web of Science databases (https://www.webofscience.com/wos/author/record/1523020) and/or having a CiteScore percentile in the Scopus database (H-index 14, https://www.scopus.com/authid/detail.uri?authorId=57197860996), 27 articles in 2017-2022); 41 in publications CCSES, co-author of 4 monographs, 4 scientific and methodological recommendations, databases.
- Almerekova, Senior Researcher, PhD (2018, Geobotany). Co-author of 35 scientific papers, incl. 12 articles, including 7 articles in peer-reviewed journals indexed in Web of Science (H-index=4, https://www.webofscience.com/wos/author/record/9617158) and Scopus (H-index=4, https ://www.scopus.com/authid/detail.uri?authorId=57196952712), and 7 articles in journals, CCSES MES RK.
- Amalova, MS (Geobotany), researcher, co-author of 5 articles (H-index 2, https://www.scopus.com/authid/detail.uri?authorId=57213623570) in Scopus/WoS, and 3 articles in CCSES.
- Yermagambetova, MS, PhD-student, junior researcher, has 1 article in Scopus/WoS.
- Podzorova, Bachelor, laboratory assistant at IPBB, master student of the 2nd year of al-Farabi Kazakh National University, ORCID ID 0000-0002-9521-5754.
List of publications of the project’s participants (2017-2022)
- Abugalieva S., Volkova L., Genievskaya Y., Ivaschenko A., Kotukhov Y., Sakauova G., Turuspekov Y. (2017). Taxonomic assessment of Allium species from Kazakhstan based on ITS and matK markers. BMC Plant Biology (IF=4.96, quartile-Q1; SJR=1.378, percentile-87), 17(2), 51-60. doi: 10.1186/s12870-017-1194-0;
- Turuspekov Y., Genievskaya Y., Baibulatova A., Zatybekov A., Kotuhov Y., Ishmuratova M., Imanbayeva A., Abugalieva S. (2018). Phylogenetic taxonomy of Artemisia species from Kazakhstan based on matK analyses. Proceedings of the Latvian Academy of Sciences. Section B: Natural, Exact, and Applied Sciences (Latvia) (SJR=0.168, percentile-39), 72(1), 29. https://doi.org/10.1515/prolas-2017-0068
- Almerekova S., Mukhitdinov N., Abugalieva S. (2017). Phylogenetic study of the endemic species Oxytropis almaatensis (Fabaceae) based on nuclear ribosomal DNA ITS sequences. BMC Plant Biology (IF= 4.96, quartile-Q1; SJR=1.378, percentile-87), 17(1), 1-9. https://doi.org/10.1186/s12870-017-1128-x
- Almerekova S., Shchegoleva N., Abugalieva S., Turuspekov Y. (2020). The molecular taxonomy of three endemic Central Asian species of Ranunculus(Ranunculaceae). PloS one (IF=3.788, quartile-Q2; SJR=0.990, percentile-92), 15(10), e0240121. https://doi.org/10.1371/journal.pone.024012;
- Almerekova, S., Abugalieva, S.*, Mukhitdinov, N. (2018). Taxonomic assessment of the Oxytropis species from South-East of Kazakhstan. Vavilovskii Zhurnal Genetiki i Selektsii= Vavilov Journal of Genetics and Breeding. (SJR=0.188, percentile-38), 22(2), 285-290.
- Genievskaya Y., Abugalieva S., Zhubanysheva A., Turuspekov Y. (2017). Morphological description and DNA barcoding study of sand rice (Agriophyllum squarrosum, Chenopodiaceae) collected in Kazakhstan. BMC Plant Biology (IF=4.96, quartile-Q1; SJR=1.378, percentile-87), 17(1), 1-8. https://doi.org/10.1186/s12870-017-1132-1;
- Özek G.; Schepetkin I.A.; Yermagambetova M.; Özek T.; Kirpotina L.N.; Almerekova S.S.; Abugalieva S.I.; Khlebnikov A.I.; Quinn M.T. (2021) Innate Immunomodulatory Activity of Cedrol, a Component of Essential Oils Isolated from Juniperus Molecules (IF=4.588, quartile-Q2; SJR=0.990, percentile-74), 26, 7644.
- Turuspekov Y., Abugalieva S., Ermekbayev K., Sato K. (2014). Genetic characterization of wild barley populations (Hordeum vulgare spontaneum) from Kazakhstan based on genome wide SNP analysis. Breeding Science, (IF= 2.63, quartile–Q2; SJR=0.799, percentile-74) 64(4), 399-403.
- Sumbembayev A., Abugalieva S., Danilova A., Matveyeva E., Szlachetko D. (2021). Flower morphometry of members of the genus Dactylorhiza Necker ex Nevski (Orchidaceae) from the Altai Mountains of Kazakhstan. Biodiversitas Journal of Biological Diversity (SJR=0.257, percentile-40), 22(8). https://doi.org/10.13057/biodiv/d220855
- Almerekova S., Favarisova N., Turuspekov Y., Abugalieva S.* (2020). Cross-Genera Transferability of Microsatellite Markers and Phylogenetic Assessment of Three Salsola Species from Western Kazakhstan. Proceedings of the Latvian Academy of Sciences. Section B: Natural, Exact, and Applied Sciences (Latvia) (SJR=0.168, percentile-39), 74 (5), 325-334. https://doi.org/10.2478/prolas-2020-0049.
- Amalova A., Kurmanbaeva M., Turuspekov E., Ivashchenko A., Abidkulova K (2018). Ontogenetic structure of cenopopulations Tulipa оstrowskiana Regel in Zailiyskiy Alatau. KazNU Bulletin. Ecological series, 56(3), 101-114. (in Russian)
- Favarisova, N., Kusmangazinov A., Karelova D., Yermagambetova M., Abugalieva S.I. 2020. Optimization of PCR Conditions for Agriophyllum, Haloxylon and Salsola Microsatellite Markers. International Journal of Biology and Chemistry 13 (1):57-68. https://doi.org/10.26577/ijbch.2020.v13.i1.06.
- Orazov A.E., Mukhitdinov N.M., Myrzagalieva A.B., Turuspekov Y.K., Shramko G., Kubentaev S.A. Distribution and characteristics of cenopopulations of Amygdalus ledebouriana on the territory of the Narymsky ridge // KazNU Bulletin. Biological series (IF=0.043). 2019. №1(78). P.36-45. DOI: https://doi.org/10.26577/eb-2019-1-1401 (in Russian)
- Turuspekov E.K., Abugalieva S.I., Almerekova Sh.S. Author’s certificate of entering information into the state register of rights to objects protected by copyright — to the database «Biodiversity of the flora of Kazakhstan» (№20035 from 01.09.2021),http://www.kazflora.kz/.
Results for 2022:
- Starting isolation and purification of the DNA of available populations of Tulipa species growing in Kazakhstan. At this stage of the project, DNA has been isolated from 110 samples of 6 populations of Tulipa species native to Kazakhstan.
- Starting identification and deposition of nucleotide sequences of ITS markers of the nuclear genome of species of the genus Tulipa. Depositing the nucleotide sequences of ITS markers of the nuclear genome of species of the genus Tulipa in the international database NCBI (National Center for Biotechnology Information).
- Starting to study the level of intra- and interspecific genetic diversity of populations of endemic, rare and endangered Tulipa species of Kazakhstan using SSR markers, to determine the population structure. At this stage of the project, the PCR conditions for SSR markers of tulips were optimized in terms of the primer annealing temperature, the concentration of dNTPs (deoxynucleoside triphosphates) and Mg2+ in the PCR reaction mixture. The results will be used to study the level of genetic diversity of populations of endemic, rare and endangered species of tulips.
Results for 2023:
- Work has begun on collecting Tulipa species populations, species identification, compiling and digitizing herbariums. In 2023, 18 populations were collected for 8 tulip species: Tulipa alberti Regel (3), Tulipa buhseana Boiss. (4), Tulipa behmiana Regel (2), Tulipa kolpakowskiana Regel (2), Tulipa greigii Regel (4), Tulipa kaufmanniana Regel (1), Tulipa dasystemon (Regel) Regel (1), Tulipa turkestanica Regel (1) in southern and southeastern Kazakhstan.
- Work continued on studying the genetic diversity of the chloroplast genome of endemic and rare Tulipa species in Kazakhstan using new generation genomic technologies. Work on studying the phylogenetic relationships of Tulipa species based on the genetic diversity of chloroplast genomes was continued. In 2023, two Red Book species Tulipa alberti and Tulipa kolpakowskiana were analyzed. Annotated nucleotide sequences of the chloroplast genomes of the species T. alberti and T. kolpakowskiana were deposited in the NCBI international database under the accession numbers OR456441 and OR456442, respectively.
- Work on identifying and analyzing the phylogenetic relationships of representatives of the species of the genus Tulipa in Kazakhstan was continued using DNA markers for barcoding the nuclear and chloroplast genome. In 2023, the ITS nucleotide sequences were deposited for 7 samples of the species T. alberti, T. kolpakowskiana, T. buhseana (2 samples for this species), T. greigii (2 samples for this species), T. turkestanica under the following identification numbers — OP482244, OP482245, OR346250, OR346251, OR346252, OR346253, OR346254. The nucleotide sequences of 5 samples of tulip species T. alberti, T. kolpakowskiana, T. greigii, T. turkestanica and T. buhseana were deposited using the matK marker under the following registration numbers: OR487142, OR487143, OR487144, OR487145 and OR487146, respectively. The rbcL marker was used to upload 5 samples of tulip species from Kazakhstan: T. kolpakowskiana, T. turkestanica, T. buhseana, T. greigii and T. ostrowskiana, to the NCBI international database under the following registration numbers: OR353397, OR353394, OR353395, OR353396 and OR353398, respectively.
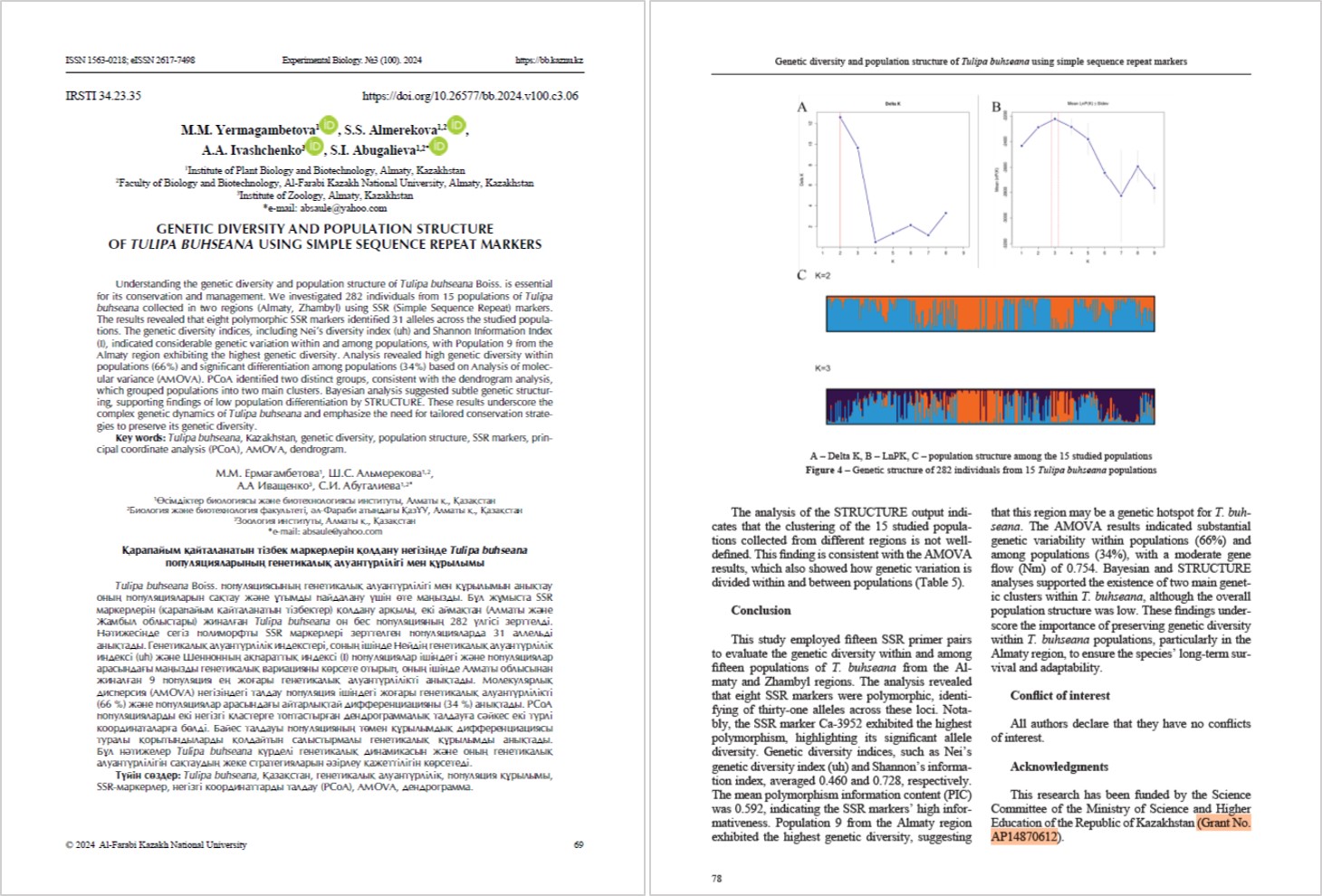
- Work continued on studying the level of intra- and interspecific genetic diversity of populations of endemic, rare and endangered Tulipa species of Kazakhstan using SSR markers to determine the structure of populations. In 2023, genotyping of 4 populations of T. buhseana, 2 populations of T. alberti and a population of T. ostrowskiana was carried out using 15 SSR markers (Ca-2572, Ca-3952, Ca-5526, Ca-5553, Ca-6950, Ca-7862, Ca-8508, Ca-13333, Ca-15730, Kn-834, Kn-1412, Kn-2291, Kn-7108, Kn-7480, Kn-30956).
- Work has begun on a phytochemical study of widespread tulip species based on secondary metabolites. Based on literature data, phenolic compounds, in particular flavonoids (coumarin, tricine, isorhamnetin, rhamnetin, kaempferol and quercetin) and organic acids (vanilla and syringic acids) were selected as the substances to be determined. Optimization of secondary metabolite extraction for determining substances on a high-performance liquid chromatograph (HPLC) was carried out.
- Work has begun on replenishing the database «Biodiversity of the Flora of Kazakhstan» with data on botanical collections, molecular genetic data of deposited nucleotide sequences of chloroplast genomes and DNA barcoding data of species of the genus Tulipa.
Results for 2024:
- The work on collecting tulip species populations, species identification, compiling and digitizing herbariums has been completed. A total of 50 populations (864 samples) of 18 tulip species were collected in the regions of Eastern, Western, Central, Southern and South-Eastern Kazakhstan. The species affiliation was determined and the compiled herbarium sheets of tulip species were digitized.
- An analysis of the complete nucleotide sequences of the chloroplast genomes of the species T. greigii, T. behmiana, T. brachystemon, T. lemmersii, T. ostrowskiana, T. tetraphylla and T. zenaidae Baker collected in Kazakhstan was carried out. The nucleotide sequences of the plastomes of the species T. greigii, T. behmiana, T. brachystemon, T. lemmersii, T. ostrowskiana, T. tetraphylla and T. zenaidae obtained in this study were deposited in the NCBI international database under the following accession numbers PP338772, PP933987, PP061001, PP061002, PP933988, PP933989 and PP061003, respectively.
- The analysis of phylogenetic relationships of the species of the genus Tulipa was carried out based on the use of DNA markers for barcoding the nuclear and chloroplast genome. The nucleotide sequences of the ITS, matK and rbcL markers of the Kazakh tulip species were used to construct the phylogenetic tree. The ITS nucleotide sequences of T. kolpakowskiana, T. lemmersii, T. zenaidae, and T. tetraphylla were deposited in the NCBI database under accession numbers PQ269144, PQ269145, PQ269146, and PQ269147, respectively. The matK sequences were deposited for five accessions of five tulip species: T. buhseana (PQ272991), T. alberti (PQ272992), T. greigii (PQ272993), T. lemmersii (PQ272994), and T. zenaidae (PQ272995). Nucleotide sequences of 6 tulip species were deposited using the rbcL marker: T. alberti (PQ272996), T. brachystemon (PQ272997), T. lemmersii (PQ272998), T. zenaidae (PQ272999), T. behmiana (PQ273000), and T. tetraphylla (PQ273001).
- Microsatellite analysis of T. alberti, T. greigii, and T. buhseana populations was performed using 15 SSR markers. Polymorphism was observed for 9 markers in 11 T. alberti populations, for 13 markers in 12 T. greigii populations, and for 8 markers in 5 T. buhseana populations. The degree and structure of genetic diversity of tulip populations in Kazakhstan was investigated using polymorphic microsatellite DNA markers. The results obtained in this project can be used to develop conservation strategies for these species in Kazakhstan.
- The work on phytochemical study of widespread tulip species for secondary metabolites has been completed. Extraction was carried out in triplicate from leaves (2 g) of T. greigii and T. kaufmanniana from the Turkestan region. The extract from T. kaufmanniana leaves contained 0.040% coumarin, 91.089% rhamnetin, 0.283% vanillic acid and 0.290% syringic acid. 117.639% vanillic acid was found in the extract from T. greigii leaves. The data obtained will be used for further study of the biological activity of these compounds. The obtained data from phytochemical analysis will contribute to the study of the chemosystematics of tulip species.
- The work on replenishing the database «Biodiversity of the Flora of Kazakhstan» with data on botanical collections, molecular genetic data of deposited nucleotide sequences of chloroplast genomes and DNA barcoding data of species of the genus Tulipa has been completed. The database «Biodiversity of the Flora of Kazakhstan» was replenished with information on botanical collections, molecular genetic data of deposited nucleotide sequences of chloroplast genome markers and the results of DNA barcoding of species of the genus Tulipa. Information on botanical collections includes data on the life form, general botanical description, ecology, distribution of species, collection sites, as well as photographs in the natural environment and herbarium specimens. In addition, links to sources are provided. Information on molecular genetic data is presented in the form of nucleotide sequences of chloroplast genome markers deposited in the international NCBI database. The database was supplemented with the above information for the species T. alberti, T. greigii, T. buhseana, T. behmiana, T. heterophylla, T. regelii, T. ostrowskiana, and T. kolpakowskiana.
Publications 2024:
List of publications in peer-reviewed foreign scientific journals indexed in the Web of Science and (or) Scopus databases
- Almerekova S., Yermagambetova M., Ivaschenko A., Turuspekov Y., Abugalieva S. Comparative Analysis of Plastome Sequences of Seven Tulipa (Liliaceae Juss.) Species from Section Kolpakowskianae Raamsd. Ex Zonn and Veldk. // International Journal of Molecular Sciences. – 2024. – Vol. 25(14). – P. 7874. https://doi.org/10.3390/ijms25147874 (IF= 5.6, Q1 Biochemistry and Molecular Biology; SJR= 1.179, percentile: 90 — Biochemistry, Genetics and Molecular Biology)
- Yermagambetova, M.; Almerekova, S.; Ivashchenko, A.; Turuspekov, Y.; Abugalieva, S. Genetic Diversity of Tulipa alberti and greigii Populations from Kazakhstan Based on Application of Expressed Sequence Tag Simple Sequence Repeat Markers // Plants. – 2024. – Vol.13. – P. 2667. https://doi.org/10.3390/plants13182667 (IF= 4.4, Q1 Plant Sciences; SJR= 0.795, percentile: 86 — Agricultural and Biological Sciences).
- Almerekova S., Yermagambetova M., Ivaschenko A., Abugalieva S., Turuspekov Y. Assessment of complete plastid genome sequences of Tulipa alberti Regel and Tulipa greigii Regel species from Kazakhstan// Genes. – 2024. – 15(11). – P.1447. https://doi.org/10.3390/genes15111447 (IF= 3.3, Q2 Plant Sciences; SJR= 0.817, percentile: 53 — Genetics)
Articles in a peer-reviewed foreign or domestic publication recommended by CQASHE:
Yermagambetova M.M., Almerekova S.S., Ivaschenko A.A., Abugalieva S.I. Genetic diversity and population structure of Tulipa buhseana using simple sequence repeat markers // Experimental Biology. No. 3 (100) – 2024. – P. 69-82. https://doi.org/10.26577/bb.2024.v100.c3.06
Catalog
Turuspekov Y.K., Ishmuratova M.Y., Almerekova S.S., Yermagambetova M.M., Abugalieva S.I. «Catalog of endemic, rare, endangered and wild economically valuable plant species of Kazakhstan. 3. Central Kazakhstan» – Almaty: Piramida Group, 2024. – 172 p.
Abstracts from international conferences:
- Almerekova S., Yermagambetova M., Turuspekov Y., Abugalieva S. Comparative analysis of complete plastome sequences of Tulipa (Liliaceae Juss.) species from Kazakhstan // XX International Botanical Congress Madrid 2024: Collection of oral presentations. – Madrid: Spain, 2024 – P. 486. (oral presentation). (In English)
- Abugalieva S., Almerekova Sh., Yermagambetova M., Turuspekov Y. Genetic divesity of wild plant species of Kazakhstan // «International Conference on Plant Biology and Biotechnology (ICPBB 2024)»: Proc. int. conf. / edited by E.K. Turuspekov, S.I. Abugalieva. — Almaty: IPBB, 2024 — 19 p. (oral presentation). (In English)
- Almerekova S., Yermagambetova M., Ivaschenko A. A., Turuspekov Y., Abugalieva S. The comparative assessment of complete plastic sequences of Tulipa (Liliaceae Juss.) species from Kazakhstan // «International Conference on Plant Biology and Biotechnology (ICPBB 2024)»: Proc. int. conf. / edited by E.K. Turuspekov, S.I. Abugalieva. — Almaty: ICPBB, 2024 — 20 p. (oral presentation). (In English)
- Almerekova S., Yermagambetova M., Ivaschenko AA, Turuspekov Y., Abugalieva S. Plastid genome sequencing of Tulipa species from Kazakhstan: insights into genomic structure, repeats, nucleotide diversity, and phylogenetic relationships. Proc. Int. Conf. «Astana Biotech 2024». Symposium «Regional Flora of Kazakhstan: Diversity, Genetics, and Conservation». September 12-13, 2024, Astana, NCB, Kazakhstan. (oral presentation). (In English)
- Abugalieva S., Almerekova Sh., Yermagambetova M., Turuspekov Y. Genetic diversity and digital documentation of Kazakhstan’s flora. Proc. Int. Conf. «Astana Biotech 2024». Symposium «Regional Flora of Kazakhstan: Diversity, Genetics and Conservation». September 12-13, 2024, Astana, NCB, Kazakhstan. (oral presentation). (In English)
Conference papers: (indicating the paper format)
- Almerekova S.S. – oral presentation at XX International Botanical Congress, Madrid, Spain, 2024. (In English)
- Abugalieva S.I. – oral presentation at the international practical conference “International Conference on Plant Biology and Biotechnology (ICPBB 2024)”, Almaty, Kazakhstan, 2024. (In English)
- Almerekova S.S. – oral presentation at the international practical conference “International Conference on Plant Biology and Biotechnology (ICPBB 2024)”, Almaty, Kazakhstan, 2024. (In English)
- Abugalieva S.I. – oral presentation at the International Conference Astana Biotech 2024, Astana, Kazakhstan, 2024. (In English)
- Almerekova S.S. – oral presentation at the International Conference Astana Biotech 2024, Astana, Kazakhstan, 2024. (In English)