Brief description of the project
(2023-2025)
Project title: IRN AP 19677444 «Identification of quantitative trait loci for productivity and quality of chickpea (Cicer arietinum L.) seeds using genome-wide association study».
Relevance. Chickpea (Cicer arietinum L.) is an important crop for food security and feed production due to its high content of edible plant protein, hypoallergenic, and balanced amino acid composition. One of the urgent tasks of genetics and breeding is the search for associations between genes, DNA markers, and breeding-valuable traits using the methodology of genome-wide association study (GWAS). This project will search for loci of quantitative traits associated with the yield and quality of chickpeas, using data from whole-genome sequencing of 4972803 SNP markers (Illumina HiSeq2500) and phenotyping of 238 chickpea samples from the world collection, in the conditions of south-east Kazakhstan. As a result, a set of informative KASP (Kbioscience competitive allele-specific PCR) markers associated with the productivity and quality of chickpea seeds will be developed. High-yielding and high-quality chickpea accessions will be selected to strengthen breeding programs in Kazakhstan. An assessment of the genetic diversity of the world chickpea collection, including samples of Kazakhstan breeding, will also be given using microsatellite markers.
The goal of the project: To search for quantitative trait loci that determine the productivity and quality of seeds, based on studying the world collection of chickpeas and using the methodology of genome-wide association study.
Expected results:
- The world collection of chickpeas will be studied for economically valuable traits in the southeast of Kazakhstan. Characteristics of the collection samples will be given based on a set of morphological characteristics and productivity components;
- The genetic diversity of the chickpea collection will be assessed using microsatellite DNA markers. The most informative DNA markers associated with economically valuable traits will be identified;
- An assessment of the biochemical composition of seeds in the chickpea collection will be given. The level of variability of protein and amino acid content in chickpea collection will be studied;
- Associations of DNA markers with productivity and quality traits of chickpea seeds will be identified using a genome-wide association study;
- Informative KASP markers for seed productivity and quality will be developed. Local commercial chickpea cultivars and promising lines will be genotyped using KASP markers;
- Validation of the effectiveness of informative KASP markers will be carried out based on the study of local hybrid lines of the older generation. Promising chickpea lines will be selected for breeding intensification in Kazakhstan.
Scientific Supervisor of the project: Anuarbek Shynar Nurlankyzy, PhD, Researcher at the Laboratory of Molecular Genetics of the IPBB, 11 years of experience on the project topic, H index – 4,https://www.scopus.com/authid/detail.uri?authorId=57192177334
Research group:
Turuspekov Yerlan Kenesbekovich, Candidate of Biological Sciences, Professor, Head of the Laboratory of Molecular Genetics. H-index 16, https://www.scopus.com/authid/detail.uri?authorId=57197860996
Abugalieva Saule Izteleuovna, Principal Researcher, Doctor of Biological Sciences, Professor. H-index 12, https://www.scopus.com/authid/detail.uri?authorId=6507029042).
Zatybekov Alibek Kamzabekovich, PhD, a leading researcher at the Laboratory of Molecular Genetics, Institute of the IPBB, H-index – 4, https://www2.scopus.com/hirsch/author.uri?accessor=authorProfile&auidList=57196942983),
Baytarakova Kuralai Zhumanovna, Master, 1st-year doctoral student at Kazakh Agrarian University, senior researcher at the Department of Leguminous Crops of the Kazakh Research Institute of Agriculture and Plant Growing.
List of publications of the project’s participants (2018-2022)
- Utility model patent KZ No. 3137 A01N 1/04 (2006.01) State Register bulletin No. 36 dated 09.24.2018. Zatybekov A.K., Anuarbek S.N., Abugalieva S.I., Doszhanova B.N., Turuspekov Y.K. A method for identification of breeding valuable soybean lines using KASP technology. https://gosreestr.kazpatent.kz/Utilitymodel/Details?docNumber=283866.
- Genievskaya Y., Pecchioni N., Laidò G., Anuarbek S., Rsaliyev A.,Chudinov, V.; Zatybekov A., Turuspekov Y., Abugalieva S., Genome‐Wide Association Study of Leaf Rust and Stem Rust Seedling and Adult Resistances in Tetraploid Wheat Accessions Harvested in Kazakhstan. Plants (IF=4.658, Q1-Plant Sciences; SJR=0.765, percentile 71 – Plant Sciences) 2022, 11, 1904. https://doi.org/10.3390/plants11151904. (https://www.scopus.com/record/display.uri?eid=2-s2.0-85136983036&origin=resultslist&sort=plf-f).
- Anuarbek S., Abugalieva S., Pecchioni N., Laidò G., Maccaferri M., Tuberosa R., Turuspekov Y. (2020). Quantitative trait loci for agronomic traits in tetraploid wheat for enhancing grain yield in Kazakhstan environments. PLOS ONE (IF=3.752, Q2- Multidisciplinary Sciences; SJR=0.852, percentile 87 – Multidisciplinary), 15(6), e0234863. doi:10.1371/journal.pone.0234863. (https://www.scopus.com/record/display.uri?eid=2-s2.0-85087025039&origin=resultslist&sort=plf-f).
- Zatybekov A., Anuarbek S., Abugalieva S., Turuspekov Y. Phenotypic and genetic variability of a tetraploid wheat collection grown in Kazakhstan. Vavilovskii Zhurnal Genetiki i Selektsii = Vavilov Journal of Genetics and Breeding. (Q3 – Agricultural and Biological Sciences; SJR=0.188, percentile 42 – General Agricultural and Biological Sciences). 2020;24(6):605-612. DOI 10.18699/VJ20.654
- Anuarbek S., Abugalieva S., Turuspekov Y. Validation of bread wheat KASP markers in durum lines in Kazakhstan. Proceedings of the Latvian Academy of Sciences, Section B: Natural, Exact, and Applied Sciences (SJR=0.143, percentile 29-Multidisciplinary). 2019. 73(5): P.462-465. DOI: 10.2478/prolas-2019-0071. (https://www.scopus.com/record/display.uri?eid=2-s2.0-85079387457&origin=resultslist&sort=plf-f).
- Utility model patent KZ No. 5061 C12Q 1/6806. State Register Bulletin No. 40 dated 10.09.2020. Turuspekov Y.K., Abugalieva S.I., Anuarbek S.N. A method for identification of breeding-valuable durum wheat lines ( durum Desf.) using molecular markers based on KASP technology. https://gosreestr.kazpatent.kz/Utilitymodel/Details?docNumber=323771.
- Zatybekov A., Abugalieva S., Didorenko S., Rsaliyev A., Turuspekov Y. (2018). GWAS of a soybean breeding collection from Southeast and South Kazakhstan for resistance to fungal diseases. Vavilovskii Zhurnal Genetiki i Selektsii. (Q3 – Agricultural and Biological Sciences; SJR=0.188, percentile 42 – General Agricultural and Biological Sciences). 22(5):536-543. DOI: https://doi.org/10.18699/VJ18.392. (https://www.scopus.com/record/display.uri?eid=2-s2.0-85052138239&origin=resultslist&sort=plf-f).
- Zatybekov A., Turuspekov Y., Doszhanova B., Didorenko S., Abugalieva S. Effect of Population Size on Genome-Wide Association Study of Agronomic Traits in Soybean // Proceedings of the Latvian Academy of Sciences, Section B: Natural, Exact, and Applied Sciences, (SJR=0.143, percentile 29-Multidisciplinary), 2020, 74(4), P. 244–251. DOI: https://doi.org/10.2478/prolas-2020-0039.
- Baytarakova K.Z., Kudaibergenov M.S., Kenenbaev S.B. Productivity and quality of chickpea grain at the final stage of breeding // Izdenister, natizheler, No. 4 (88) 2020. – P. 199-204 [In Russian].
- Patent for breeding achievement KZ No. 943. State Register Bulletin No. 53 dated 31.12.2020. Baytarakova K.Z., Begzhanov Z.N., Kudaibergenov M.S., Aldeshova M.K. Chickpea cultivar «Nurly-80». Copyright certificate No. 598 dated March 13, 2017(https://gosreestr.kazpatent.kz/SelectionAchievement/Details?docNumber=291390).
- Patent for breeding achievement KZ No. 943. State Register Bulletin No. 53 dated 31.12.2020. Bulatova K.M., Mazkirat Sh., Orazaliev N.N., Kudaibergenov M.S., Baytarakova Z. Chickpea cultivar «Miras 07».(https://gosreestr.kazpatent.kz/SelectionAchievement/Details?docNumber=282272).
- Zatybekov A., Genievskaya Y.; Rsaliyev A., Maulenbay A., Yskakova G., Savin T., Turuspekov Y., Abugalieva S. Identification of Quantitative Trait Loci for Leaf Rust and Stem Rust Seedling Resistance in Bread Wheat Using a Genome-Wide Association Study. Plants (IF=4.658, Q1- Plant Sciences; SJR=0.765, percentile 71 – Plant Sciences) 2022, 11, 74. https://doi.org/10.3390/plants11010074. (https://www.scopus.com/record/display.uri?eid=2-s2.0-85121687724&origin=resultslist&sort=plf-f).
Results for 2023: As a result of field tests of 238 samples of the world chickpea collection in the Almaty region, a comparative assessment of growing seasons and yield components of the studied samples was conducted. Samples with the highest trait performance based on the weight of seeds per plant (8 accessions) and the 1000 seed weight (4 accessions) were identified. The extraction and purification of DNA of chickpea were carried out in 3 replications. Primer screening of 5 SSR markers was carried out.
Publications (2023): ————
Results for 2024:
The study of the world collection of chickpeas on economically valuable characteristics in the south-east of Kazakhstan was continued. The phenotypic variability of the samples was assessed based on a structural analysis of a complex of morphological characteristics and productivity components. The assessment of the genetic diversity of the chickpea collection continued using 12 microsatellite DNA markers associated with economically valuable traits. The assessment of the biochemical composition of the chickpea collection seeds continued. The study of the level of variability in the chickpea collection in terms of protein and amino acid content have been continued. In this chickpea collection, the protein content ranged from 13.5% to 37.9%, with an average value of 27.0±4.9 %. 35% of the collection was characterized by high protein content (30-38%), while the majority of the collection (60%) contained protein in the range of 20-29%. The identification of associations between DNA markers and productivity and quality traits of chickpea seeds has begun based on the use of genome-wide association analysis methodology. 41 quantitative trait loci (QTLs) were identified on 7 chickpea chromosomes. Based on yield components (number of grains per plant, weight of 1000 grains, weight of grains per plant), 30 LCPs were identified on chromosomes 1, 2, 3, 4, 6, 7 and 8. Based on adaptability components (plant height, height of attachment of the lower bean), 3 LCPs were identified on chromosomes 2, 4 and 6. Based on phenological phases (VER2, VER8, R2R4), 8 LCPs were identified on chromosomes 1, 2, 4. The creation of informative KASP markers for traits of productivity and seed quality has begun. Genotyping of local commercial varieties and promising chickpea lines has begun using the created KASP markers. Validation of the effectiveness of informative KASP markers has begun based on the study of local hybrid lines of the older generation.
Results for 2025
Evaluation of 258 global chickpea (Cicer arietinum L.) genotypes revealed significant differences in agronomic traits depending on their region of origin. Overall, across the combined sample from all origin groups, the average plant height was 33.68 cm, the height of the lower pod attachment was 18.63 cm, the average number of lateral branches per plant was 2.23, and the yield per plant was 4.71 g. These results highlight the high breeding potential of domestic accessions (Figure 1).
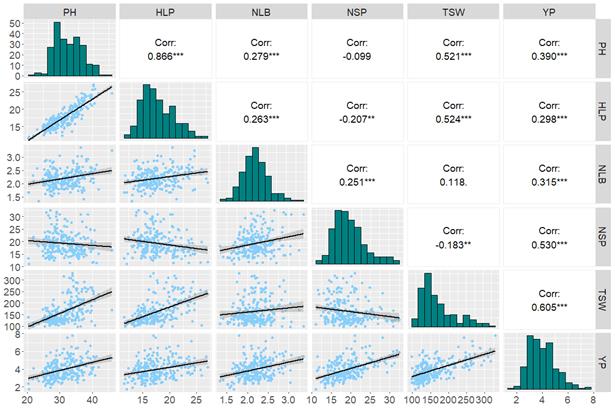
PH – plant height; HLP – height of lower pod; NLB – number of lateral branches; NSP – number of seeds per plant; YP – yield per plant; TSW – 1000-seed weight
Figure 1 – Correlation analysis of traits valuable for breeding
Analysis of variance revealed a significant effect of the factors “origin,” “seed type,” and “year of study,” as well as their interactions, on several breeding-relevant traits in the 258 chickpea genotypes studied.
A total of 28 SSR markers were used for genotyping the Kazakhstan chickpea collection. Of these, 23 markers were polymorphic, revealing 2 to 8 alleles per locus, while five markers were monomorphic. Combined results obtained from STRUCTURE, PCA, and pairwise genetic distance analyses indicated the presence of four main genetic clusters among 27 chickpea accessions, with a high degree of admixture and no clear genetic differentiation between desi and kabuli types, reflecting the shared and complex genetic background of the studied collection (Figure 2).
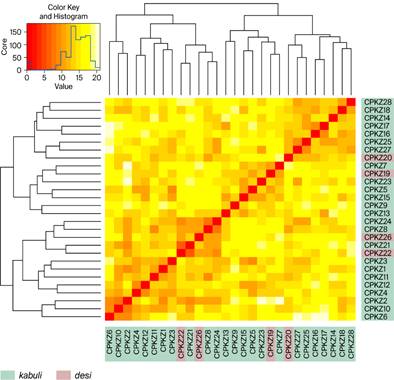
Figure 2 – Genetic relationships between 27 chickpea accessions from Kazakhstan based on SSR markers
Phenotypic evaluation of the biochemical composition of seeds from 258 chickpea accessions revealed considerable polymorphism in key nutritional traits, highlighting the high suitability of this collection for breeding programs. Principal component analysis (PCA) showed that PC1 explained 44.3% of the total variance and reflected a pronounced negative correlation between protein content and both fat and moisture content, whereas PC2 (31.5% of variance) captured variability in the amino acid profile, associated with the contrast between asparagine and glutamine. Together, these two components accounted for 75.8% of the total data variability (Figure 3).
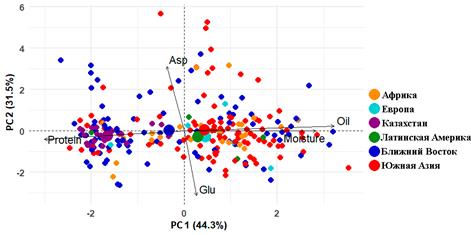
Figure 3 – Principal component analysis showing the distribution of accessions from different regions of the World by the studied traits. Asp – aspartic acid; Glu – glutamic acid
A genome-wide association analysis (GWAS) was conducted to identify 351 significant marker–trait associations (MTAs) for six key agronomic traits. Across the years, 128, 164, and 102 significant MTAs were identified in 2022, 2023, and 2024, respectively, with five associations remaining significant across multiple years, indicating the presence of stable genomic regions. Considering linkage disequilibrium (LD) decay, a total of 128 quantitative trait loci (QTLs) associated with the six traits were identified (65, 59, and 54 QTLs per year), of which 40 were stable, including 10 QTLs significant across all three years (Figure 4). Analysis of the phenotypic variance explained (R²) revealed varying effect sizes, ranging from low and moderate effects reflecting the polygenic nature of the traits to seven QTLs with high R² (>0.10), corresponding to the major loci Q_NLB_1.1, Q_NLB_1.2, Q_NSP_4.2, Q_TSW_4.1, Q_TSW_4.2, Q_YP_1.1, and Q_YP_2.1.
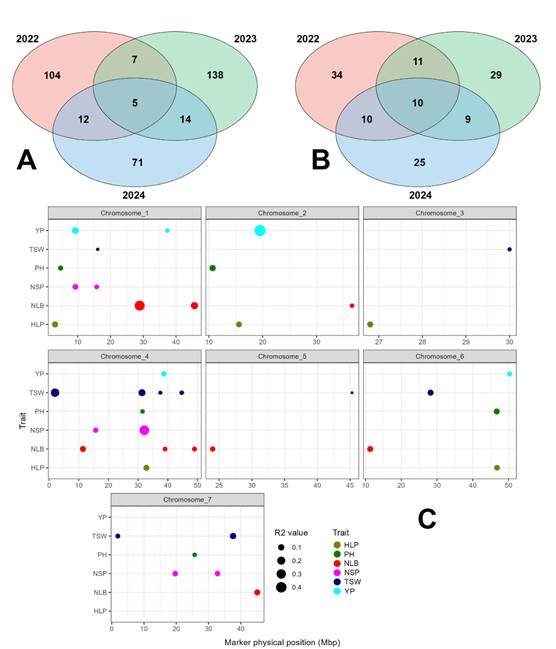
Total MTAs by year (A), stable QTL by year (B) and the effect (R2) of identified markers on chromosomes (C)
Figure 4 – GWAS results with distribution of significant marker-trait associations (MTAs)
Analysis of the distribution of QTLs across the genome showed that the majority (70.3%) were located in intergenic regions, while the remaining 29.7% were within coding sequences. SNP markers identified through GWAS were subsequently converted into more user-friendly KASP markers. As a result, 27 SNP markers were successfully converted into highly specific and cost-effective KASP markers associated with agronomically important traits.
In the next step, a collection of 38 accessions was subjected to KASP genotyping. Of the 19 KASP markers analyzed, 17 (89%) showed statistically significant associations with one or more phenotypic traits, indicating their high informativeness and potential value for further use in breeding programs. The identified KASP markers, in particular ipbb_ca_005, ipbb_ca_008, ipbb_ca_013, and ipbb_ca_023 (yield-related components), and ipbb_ca_003, ipbb_ca_021, and ipbb_ca_026 (morphological traits), can be considered reliable candidates for use in marker-assisted selection programs for chickpea aimed at improving yield and optimizing plant morphological characteristics.
Information for potential users
The study of a chickpea (Cicer arietinum L.) collection, including 258 global genotypes and Kazakhstan accessions, has high practical significance for breeding, seed production, and sustainable crop production in the Republic of Kazakhstan. Observed differences in key agronomic traits and seed quality parameters, as well as the high level of genetic diversity (four genetic clusters with substantial admixture), confirm the high breeding potential of the studied material and its suitability for targeted use in breeding programs across different agro-ecological zones of the country.
Within the framework of GWAS, significant marker–trait associations and key quantitative trait loci, including stable and high-effect loci, were identified, providing a scientific basis for marker-assisted selection. Conversion of SNPs into informative KASP markers and their successful validation support the recommendation of these results for practical implementation, aimed at accelerating the development of competitive, high-yielding, and adapted chickpea varieties, thereby enhancing crop productivity and strengthening food security in the Republic of Kazakhstan.
Publications 2025
List of publications in peer-reviewed scientific journals indexed in the Web of Science and (or) Scopus databases
Zatybekov A., Genievskaya Y., Anuarbek,S., Kudaibergenov M., Turuspekov Y., Abugalieva S. Phenotypic and Genetic Diversity of Chickpea (Cicer arietinum L.) Accessions from Kazakhstan // Diversity. – 2025. – Vol.17, – P.664. https:// doi.org/10.3390/d17090664.
Articles in a peer-reviewed domestic publication with a non-zero impact factor (recommended by the SHEQAC MSHE RK)
Zatybekov A.K., Yeshengaliyeva A.N., Anuarbek Sh.N., Kudaibergenov M.S., Turuspekov Y.K., Abugalieva S.A. Field Evaluation and Diversity of 238 Global Chickpea (Cicer arietinum L.) Genotypes Grown in South-East Kazakhstan // Fundamental And Experimental Biology. – 2025. – Vol.30. – P.3(119).
Abstracts of the international conference (indicating the form of the report )
Zatybekov A.K., Burakhoja A.K., Kudaybergenov M.S., Varshney R., Krempa A., Anuarbek Sh., Turuspekov Y.K. Variability of agronomic traits in chickpea collection grown in irrigated and non-irrigated conditions // International Conference on Plant Biology and Biotechnology (ICPBB 2024). – Almaty, 2024. – P. 111 (постер).

