Brief description of the project
(2023-2025)
Project title: AP19678215 «Metagenomic and population analysis of potato viruses and viroids and study of the mechanisms of their interaction with resistant potato varieties and hybrids».
Relevance.
This project is aimed at solving problems related to controlling the spread of potato viral infections, preventing the importation of dangerous strains and isolates of economically important viruses and viroids from other countries, and qualitatively increasing the breeding potential of varieties resistant to virus infection. Intensification of potato breeding in the country through cultivation of resistant local potato varieties and control over the spread of viruses and their vectors will make it possible to come to import substitution and dependence on foreign seed material. Molecular and genetic study of potato viruses and their interaction with potato varieties of local selection will prevent the spread of viruses in potato fields, especially seed farms.
The goal of the project:
The goal of the project is to investigate potato virome, study the genetic diversity of potato viruses and viroids distributed in Kazakhstan, as well as the mechanisms of interaction between natural isolates of viruses and viroids and resistant potato varieties, and identify new genes associated with the immune response to viral infection.
Expected results:
1) At least 500 potato samples will be collected from potato growing regions of the country (Northern and Almaty regions). Metagenomic analysis of potato virome, species identification of viruses and viroids common in potato growing fields of the country. Detection of new, quarantine and particularly dangerous viruses and viroids. Tubers and above-ground parts will be collected from plants showing signs of infestation as well as from asymptomatic plants. Tubers will be used as a source of virions for inoculation of resistant plants.
2) Full genome sequencing of detected PLRV, X, Y, S and M viruses and PSTVd potato viroid causing the greatest economic damage, detection of isolates and strains circulating in the country will be conducted. Development of a new vir-typing method for viruses to determine the genetic diversity of isolates in geographically distant populations. Comparison of the results of population analysis by vir-typing and full-genome sequencing. The new typing method will manifold reduce the cost of studying the population genetics of viruses. Identification of natural strains/isolates with significant mutations in the proteins responsible for amplification, movement and suppression of RNA interference, revealing the impact of mutations on virus biology by inoculation. Identification of the level of intraspecific and interspecific recombination. Identification of new recombinant forms. Whole-genome sequencing will identify conserved and variable patterns affecting the infectivity of the virus.
3) Identification of introgressed resistance genes to economically important potato viruses and viroids in 250 potato varieties and hybrids of Solanum tuberosum of local and foreign selection will be conducted. The selection of local potato varieties and hybrids involves a variety of source lines, which leads to a positive probability of identifying resistant alleles, as we have shown in the study of these varieties for the presence of resistance alleles to X and A potato viruses.
4) Selected potato varieties will be inoculated with different combinations of mutants in genes encoding proteins responsible for amplification, movement and suppression of RNA interference. Determination of the level of amplification and systemic movement of different virus isolates in plants carrying resistance genes. Determination of the relationship of activity of resistance genes in a specific genetic environment (different cultivars) to selected virus isolates. Detection of the influence of genetic environment on the activity of resistance genes. Identification of pathogenicity of selected isolates/ strains.
5) Transcriptomic analysis of potato varieties inoculated with virus isolates carrying significant mutations in target genes, identification of new genes involved in potato resistance to viral pathogens, including when inoculated with several viruses, will be conducted.
6) A bank of samples of new viral pathogens, strains and isolates will be created. Development of methods for breeders with description of genetic passports for resistance genes to economically important viruses and degree of resistance for each variety.
Scientific Supervisor of the project:
Dilyara Gritsenko
Research group:
Gritsenko D.A. — Head of Laboratory
Nizamdinova G.K. — senior researcher
Pozharsky A.S. — researcher
Taskuzhina A.K. — junior researcher
Kapytina A.I. — junior researcher
Kerimbek N.M- junior researcher
Kostyukova V.S.- junior researcher
Kolchenko M.V. — laboratory assistant
Abdrakhmanova A.B. — laboratory assistant
Adilbaeva K. — laboratory assistant
A.N.Makhambetov — laboratory assistant
List of publications of the project’s participants (2018-2022)
- Gritsenko, D., et al. Development of a “deconstructed” vector based on the genome of grapevine virus A // Plant Biotechnol Rep. -2019. Индекс цитирования – 2, Процентиль – 64, Квартиль- Q2, DOI: 10.1007/s11816-019-00528-1.
- Pozharskiy, A., Kostyukova, V., Nizamdinova, G., Kalendar, R., & Gritsenko, D. (2022). MLO proteins from tomato (Solanum lycopersicum L.) and related species in the broad phylogenetic context. Plants, 11(12), 1588.. DOI: 10.3390/plants11121588; WOS. Q1 (IF 4,827), Scopus: процентиль 71.
- Gritsenko, D., Pozharskiy, A., Dolgikh, S., Aubakirova, K., Kenzhebekova, R., Galiakparov, N., Sadykov, S. Apple varieties from Kazakhstan and their relation to foreign cultivars assessed with RosBREED 10K SNP array. 2022. Eur.J.Hortic.Sci. 87 (1) 1-8, DOI: 10.17660/eJHS.2022/006. Индекс цитирования – 0, Процентиль – 62, Квартиль- Q2.
- Pozharskiy, A., Kostyukova, V., Taskuzhina, A., Nizamdinova, G., Kisselyova, N., Kalendar, R., Gritsenko, D. (2022). Screening a collection of local and foreign varieties of Solanum lycopersicum L. in Kazakhstan for genetic markers of resistance against three tomato viruses. Heliyon. DOI: 10.1016/j.heliyon.2022.e10095; WOS. Q2 (IF-3.7), Scopus: процентиль 82.
- Gritsenko, D., Zulfiya Kachiyeva, Gulzhan Zhamanbayev, Bakhytzhan DuisembekoV, Abai Sagitov. Detection of five potato viruses in Kazakhstan // IX International scientific agriculture symposium “AGROSYM 2018”., p. 611. 2018.
- Gritsenko D., Aubakirova K., Galiakrapov N. Simultaneous detection of five apple viruses by RT-PCR. International Journal of Biology and Chemistry (2020) v. 13, n. 1, p. 129-134. 2020. Индекс цитирования –0, doi: 10.26577/ijbch.2020.v13.i1.13.
- Gritsenko, D., et al. «Detection of Grapevine virus A in wild grape in Kazakhstan.» Phytopathology. Vol. 109. No. 11. 3340 Pilot Knob Road, St Paul, Mn 55121 Usa: Amer Phytopathological Soc, 2019.
- Gritsenko, D. A., K. P. Aubakirova, and A. S. Pozharskiy. «SSR profiling of potato cultivars resistant to pathogens.» Plant Genetics, Genomics, Bioinformatics, and Biotechnology. 2021.
- A.S. Pozharskiy, K. Aubakirova, D. Gritsenko, N. Galiakparov. Genotyping and morphometric analysis of Kazakhstani grapevine cultivars versus Asian and European cultivars // Genet. Mol. Res., 2020. Индекс цитирования – 0, DOI: 10.4238/gmr18482.
- A. Pozharskiy, D. Gritsenko // Prediction of Slmlo1 protein paralogs in Solanum l. Spp. using partially assembled genomic dat // IV. International Agricultural, Biological & Life Science Conference – 2022 – P.162.
- A. Kapytina, N. Kerimbek, A. Taskuzhina, G. Nizamdinova, K. Adilbayeva, S. Murzatayeva, Z. Kachiyeva // Detection and genetic investigation of potato leafroll virus in Kazakhstan // IV. International Agricultural, Biological & Life Science Conference – 2022 – P.165.
- Kerimbek N., Kapytina A., Pozharsky A., Khusnitdinova M., Gritsenko D. // Phylogenetic analysis of raspberry bush dwarf virus // Proceedings of the International Scientific and Practical Conference «Actual problems and prospects of science development in the field of fruit and vegetable growing» -2022 — P.139
Results for 2023:
Nucleotide sequences of the complete genome or individual regions were analyzed for PLRV in the amount of 296, PVX- 720, PVY- 632, PVS- 439, PVM- 276, PSTVd- 2864. The analysis identified at least 5 promising regions for primer design for each target pathogen. The variability within genomes ranged from 3.4 to 15.3%; within individual genes, the average variability did not exceed 4.7%. For primer design, a region was used whose conservation within species was at least 99.5% when analysing different strains and isolates; variability outside the species was at least 60%.
PLRV (11%), PVX (37%), PVY (29%) and Alfalfa mosaic virus (4%) were analyzed for the laboratory potato collection. Homology among isolates of each PLRV, PVX and PVY virus was 87 — 95% by full genomic analysis. Among PLRV isolates, the mutation frequency did not exceed 1.3 × 10-3 , for PVX 1.01 × 10-3 , for PVY virus 1.15 × 10-3 . The highest mutation frequency was observed in PLRV virus.
Metagenomic analysis of aboveground plant parts as well as potato tubers collected from the fields was performed. The analysis identified PLRV, X, Y, S viruses and viroid PSTVd. The northern region of the country is dominated by X and Y viruses, while the southern region (Narynkol) is dominated by X virus. In addition to the above viruses, Alfalfa mosaic virus was also detected.
Whole-genome analysis of PLRV, PVX and PVY viruses showed a high homology within each isolate population studied, which was 97 — 100%. Among PLRV isolates, the mutation frequency ranged from 0.95 × 10-3 to 1.19 × 10-3 , for PVX from 0.82 × 10-3 to 0.98 × 10-3 , and for PVY virus did not exceed 0.92 × 10-3.
Results for 2024:
Whole genome sequencing was conducted for 43, 48, and 11 isolates of S virus, M virus, and PSTVd viroid, with genome coverage depth from 180 to 250X, providing complete genome coverage (100%). Vir-typing resulted in 132 polymorphic amplicons for S virus, 161 for M virus, and 115 for PSTVd. M virus showed high variability both within regions (Northern and Southern) and between them, with a genetic distance of 0.124, while homology within regions did not exceed 89%. Phylogenetic analysis showed that local S virus isolates form subclades with PVSI isolates, while M virus isolates form subclades with PVM-o isolates. For X virus, genetic distance between populations (0.198) was established, with separate clades forming for each region. Local Y virus isolates formed subgroups with phylogroup I-1, and the genetic distance within the population was 0.087. The closest relationship was found with phylogroup O. For PLRV virus, the open reading frames encoding RTD and P0 showed the highest variability, with variability levels of 35.62% and 27.615% respectively, with 9 significant mutations affecting protein biological activity identified. For S and M viruses, capsid proteins were most variable, with variability levels of 21% and 19.3% respectively, while M virus showed 5 unique variants affecting protein biological activity. Y virus showed highest variability in genes encoding P1 and VPg proteins, with variability levels of 21.3% and 12.1%, but no significant mutations affecting protein biological activity were identified. For X virus, the gene encoding capsid protein showed highest variability at 19.8%, and 15 significant mutations affecting viral particle assembly and movement were identified, including 3 mutations in the presumed genome interaction region. 50 potato samples were selected for the study. The sample included varieties and hybrids with various agronomic characteristics adapted to Kazakhstan’s climatic conditions. Main varieties include both early and late-maturing varieties with varying yields and field resistance to major viral pathogens. To confirm genomic DNA quality and suitability for further analysis, quality control was performed by amplifying the 28rRNA housekeeping gene. The following markers were selected for screening genes responsible for potato virus resistance: for Y virus resistance – ADG1, ADG2, RysC3, RY186, GP122/718, GP122/564, SCARysto4; for X virus – SPUD237, CP60, PVX; for PRLV virus – RGASC850; and for PVS virus – SC811/260, CP16. Results showed that Maxim, Resource, Berkut varieties, and the Ts-6 hybrid have the highest number of loci associated with resistance to these viral pathogens. Infected potato tubers were germinated under controlled conditions in a climate chamber. For germination, samples previously tested for viral pathogens were used, with at least 10 tubers infected with each virus separately selected from each region, as well as tubers infected with multiple viruses simultaneously.
Publications and patents
l Adilbayeva, K., Moisseyev, R., Kolchenko, M., Kenzhebekova, R., Khassanov, V., Beisembina, B., … & Gritsenko, D. (2024). Genetic Evaluation of Kazakhstani Potato Germplasm for Pathogen and Pest Resistance Using DNA Markers. Agronomy, 14(9), 1923.
2 Кенжебекова, Р. Т., Мендыбаева, А. С., Капытина, А. И., & Гриценко, Д. А. (2024). Распространение смешанных вирусных инфекций картофеля в алматинской области казахстана. Вестник КазНУ. Серия биологическая, 99(2), 91-100.
Results obtained for the 2025 objectives
A transcriptomic analysis of potato cultivars infected with PVY, PVX, PVS, PLRV, and PVM at 21 dpi was performed to identify the expression of genes associated with mechanisms of resistance to viral infections. This analysis made it possible to reveal patterns of gene regulation involved in antiviral resistance mechanisms and demonstrated pronounced differences among cultivars in the level of transcriptional activity.
Based on high-throughput sequencing data, gene expression profiles were constructed, correlations between transcriptional patterns were analyzed, and functional annotation of the most highly expressed genes was carried out on average for all viruses and their combinations.
Analysis of expression profiles enabled the identification of eighteen genes with the highest transcriptional activity in the studied potato cultivars. Transcription levels varied significantly among genotypes: the highest expression values were observed in the cultivars Berkut and Zhigulevsky, whereas the cultivars Dina and Maksim exhibited minimal expression levels.
Functional analysis of the most highly expressed genes showed that the active transcripts are predominantly involved in key metabolic and regulatory processes that ensure plant adaptation to biotic stresses, including viral infection.
Correlation analysis revealed a high degree of coordinated expression among potato cultivars for the major genes involved in antiviral defense and photosynthesis.
Based on the results of molecular detection, whole-genome sequencing, and metagenomic analysis, a repository of pathogen samples was established, comprising molecularly characterized isolates of the viruses PLRV, PVX, PVY, PVS, PVM, and the viroid PSTVd.
Scientific and methodological recommendations intended for use in breeding programs were published, aimed at the molecular genetic profiling of potato cultivars, hybrids, and lines based on resistance genes to economically significant viruses, with an assessment of the resistance level of each genotype.
Information for potential users
The potential for applying the project results in both scientific research and agricultural production is substantial. The developed diagnostic test systems can be implemented in potato virus monitoring programs to enable rapid detection of emerging threats and to prevent their spread across cultivated areas. The identified potato genotypes carrying resistance genes to economically significant viral pathogens represent an important resource for targeted breeding of high-yielding and resilient cultivars. The scientific data and methodological materials generated within the project are available for use in higher education institutions of the Republic of Kazakhstan.
The effectiveness of the conducted research is confirmed by the successful completion of all project stages and the achievement of highly significant scientific results published in high-ranking peer-reviewed journals. The comprehensive genetic analysis of potato virus and viroid isolates enabled the identification of novel pathogen variants characteristic of Kazakhstan and revealed their associations with resistance levels in local and internationally bred potato cultivars. Genes involved in the formation of plant immunity to viral infections, including cases of mixed infections, were identified. The obtained results provide a solid scientific basis for the development of innovative breeding strategies and the production of virus-free planting material without direct genomic modification of potato plants, thereby enhancing the technological and economic efficiency of plant protection against viral diseases.
Patents:
- Гриценко Д.А., Керімбек Н.М., Адильбаева К.С., Дулат Б. Патент на полезную модель № 10787 (2025) «Набор синтетических олигонуклеотидов для обнаружения вируса скручивания листьев картофеля»
Methodological Recommendations:
1) Адильбаева К.С., Моисеев Р.М., Капытына А.И., Проценко Е.В., Гриценко Д.А. (2025). Молекулярно-генетическая паспортизация сортов, гибридов и линий картофеля отечественной и зарубежной селекции. — Алматы: Жебе, — 76 с. — ISBN 978-601-08-5440-6.
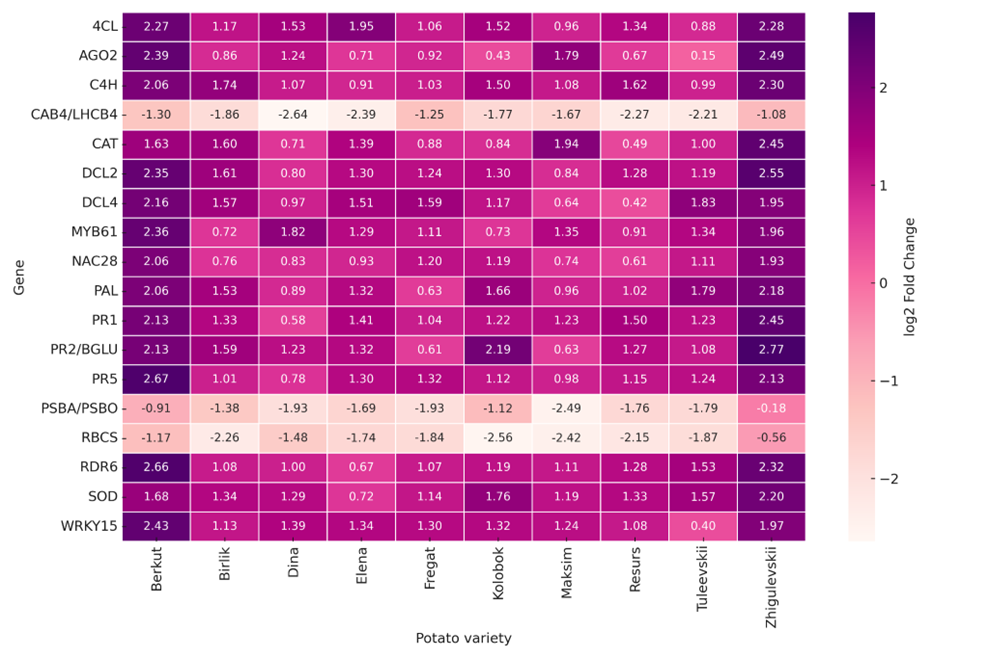
Figure 1 – Heatmap of expression levels of the most highly expressed genes in potato cultivars.
The heatmap visualizes normalized expression values (from −3 to +3) for eighteen of the most actively transcribed genes across different potato cultivars. The X-axis represents potato cultivars, while the Y-axis shows the genes with the highest expression levels. The color scale reflects transcriptional activity, where negative values indicate reduced expression and positive values indicate increased gene expression relative to the mean level.
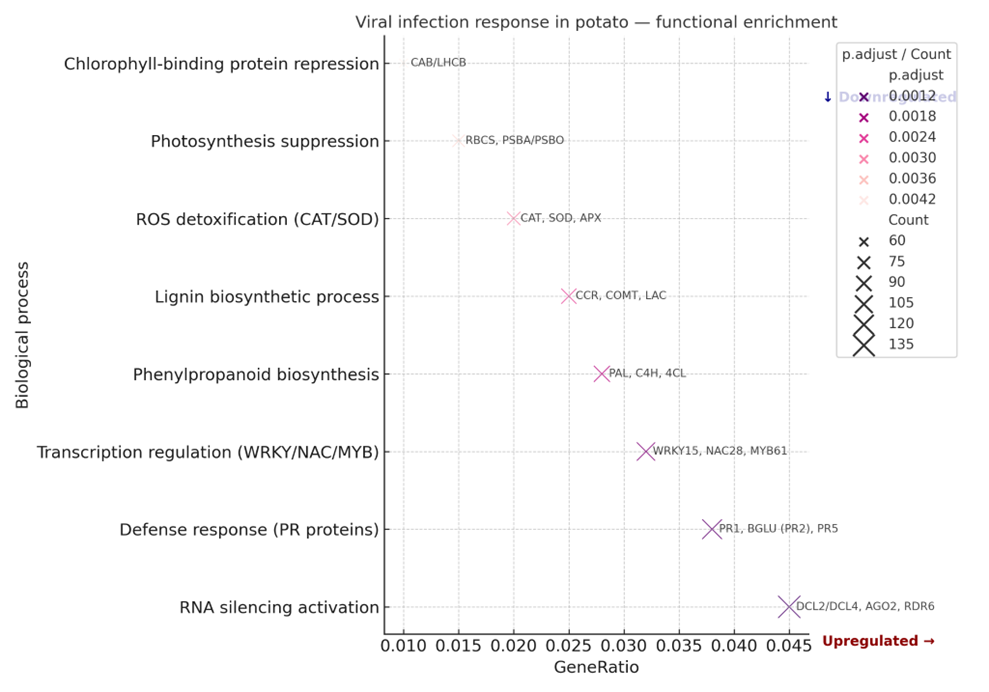
Figure 2 – Functional annotation of the most highly expressed genes (GO analysis).
The X-axis represents the GeneRatio, indicating the proportion of genes involved in each biological process, and the Y-axis shows the names of the biological processes. The color scale reflects the level of statistical significance (adjusted p-value, p.adjust), while the size of the points corresponds to the number of genes associated with each process.
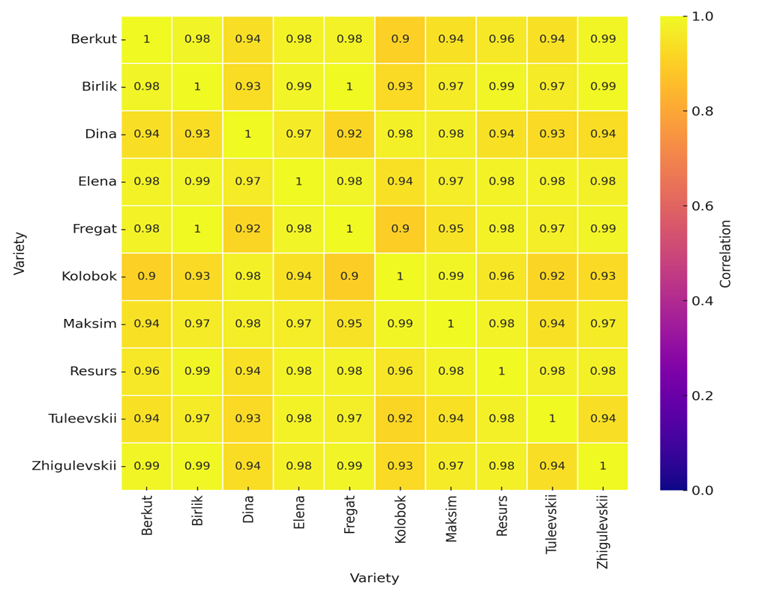
Figure 3 – Gene expression correlation matrix among potato cultivars.
The X and Y axes indicate the names of the cultivars, and the color scale represents Pearson correlation coefficients (ranging from 0 to 1). Purple shades correspond to low similarity in expression profiles, while yellow shades indicate a high degree of correlation between cultivars.

